| Version 31 (modified by , 12 years ago) ( diff ) |
|---|
Modification of the model code for the ensemble integration
Implementation Guide
Contents of this page
Overview
Numerical models are typically implemented for normal integration of some initial state. For the data assimilation with a filter algorithm, an ensemble of model states has to be integrated for limited time until observations are available and an analysis step of the filter is computed. Subsequently, the updated ensemble has to be integrated further. To allow for the interruption of the integrations by the analysis step, the model code has to be extended. As described on the page on the implementation concept of the online mode, there are two options for the ensemble integration:
- fully parallel: For this implementation one needs to use a parallel computed with a sufficient number of processes such that the data assimilation program run be run with a concurrent time stepping of all ensemble states. Thus, if one runs each mode task with n processes and the ensemble has m members, the program has to run with n times m processes. This parallelism allows for a simplified implementation as each model task integrated only one model state and the model is always going forward in time.
- flexible: This variant allows to run the assimilation program in a way so that a model task (set of processors running one model integration) can propagate several ensemble states successively. In the extreme case, this could mean that one only a a single model task that is successively performing the integration of all ensemble states. The implementation for this variant is a bit more complicated, because one has to ensure that the model can jump back in time.
The extension to the model code for both cases is depicted in the figure below (See also the page on the implementation concept of the online mode.) As at this stage of the implementation the calls to init_parallel_pdaf and init_pdaf are already inserted into the code, the difference is in the addition of routines for the time stepping.
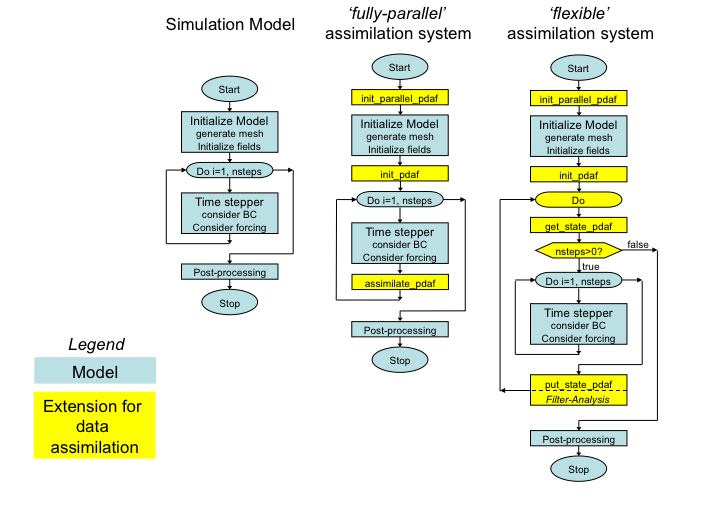
Figure 1: (left) Generic structure of a model code, (center) extension for fully-parallel data assimilation system with PDAF, (right) extension for flexible data assimilation system with PDAF.
Operations that are specific to the model and to the assimilated observations are performed by call-back routines that are supplied by the user. These are called through the defined interface of PDAF. Generally, these user-supplied routines have to provide quite elementary operations, like initializing a model state vector for PDAF from model fields or providing the vector of observations. PDAF provides examples for these routines and templates that can be used as the basis for the implementation. As only the interface of these routines is specified, the user can implement the routines like a routine of the model. Thus, the implementation of these routines should not be difficult. The implementation of the call-back routines is identical for the fully-parallel and flexible implementation variants.
The implementations that are required for the fully-parallel and the flexible implementation variants are described on separate pages:
