| Version 4 (modified by , 9 months ago) ( diff ) |
|---|
Adding an Assimilation Method to PDAF
Contents of this page
| This page describes the implementation for PDAF3. The previous implementation for PDAF2 is described on the Page on adding a DA Method to PDAF 2 |
PDAF provides an internal interface to add a data assimilation (DA) method to PDAF. Here we describe the implementation strategy and internal structure of PDAF valid for version 3.0 and later. In this text, we assume that the reader is already familiar with PDAF to the extend that it is known how PDAF is connected to a model as is described in the Implementation Guide.
The internal structure of PDAF is organized into a generic part providing the infrastructure to perform ensemble forecasts and the actual analysis step of the DA method. This generic part is independent of the particular filter algorithm. The specific routines for a DA method are called through the internal interface.
In PDAF, each DA algorithm consists of 5 Fortran modules including different subroutines for configuring the DA method, for the handling of the ensemble forecasts, and for the analysis step. The modules and routines are described below.
We provide templates for the implementation of global and local ensemble filter in the sub-directores of templates/analysis_step/.
PDAF's Internal Interface
Here, we first provide an overview of the internal interface routines of PDAF. The structure of the internal interface of PDAF is depicted in Figure 1 (For the method-specific routines, 'X' is the name of the DA method).
The left column in Fig. 1 shows the generic PDAF routines, which are called from the user code. Here, PDAF_init calls 5 interface routines to perform the specific initialization of the DA method. The other generic routines are more focused and call only one interface routine each.
The internal interface routines allowthe user to call the generic routines, which are then mapped to a specific routine of the DA method. The internal interface routines are depicted in the middle column of Fig. 1. All these routines are collected in the module PDAF_utils_filters in the file PDAF_utils_filters.F90. For each interface routine there is a specific routine of the DA method shown in the right-most column. These specific routines are collected in the module PDAF_X in the file PDAF_X.F90. The interface routines perform the initialization of a DA method, setting parameters or printing information about the configuration or the available options.
The assimilation routines, here the universal routines PDAF3_assim_offline and PDAF3_assimilate, directly call the specific assimilation routine of the DA method. These are stored the module in `PDAF_assimilate_X.F90.

Figure 1: Structure of the internal interface of PDAF. There are 7 internal interface routines (middle column) that connect the generic part with filter-specific routines. All these interface routines are collected in the module PDAF_utils filters. Each of the internal interface routines call one routine that is specific to the DA method. These routines are collected in the module PDAF_X, where 'X' would be the name of the DA method. The assimilation routines for the online and offline coupled modes are collected in the module PDAF_assimilate_X.
The separate routines are the following:
Internal interface routines
The purpose of the internal interface routines is as follows
| Interface routine in PDAF_utils_filters | called specific routine in PDAF_X | Description |
|---|---|---|
PDAF_init_filters | PDAF_X_init | Perform the filter-specific initialization of parameters and calls the user-supplied routine that initializes the initial ensemble of model states. |
PDAF_alloc_filters | PDAF_X_alloc | Allocate the filter-specific arrays. |
PDAF_options_filters | PDAF_X_options | Display an overview of available options for the filter algorithm. |
PDAF_set_iparam_filters | PDAF_X_set_iparam | Set integer parameter for the DA method |
PDAF_set_rparam_filters | PDAF_X_set_rparam | Set real (floating point) parameter for the DA method |
PDAF_print_info_filters | PDAF_x_memtime | Display information on the run time of the different parts of the DA method as well as information on the amount of allocated memory. |
PDAF_configinfo_filters | PDAF_x_config | Display the current configuration of the DA method |
When PDAF_init is called, the DA method is chosen by its ID number or its name parameter (see page on specific options of DA method). Internally to PDAF, each DA method is identified by a string that is defined in the module PDAF_DA in PDAF_da.F90. The interface routines have a very simple structure. In general, they select the method-specific routine based on the string identifying the filters.
Collecting all interface routines in the file PDAF_utils_filters.F90 yields a single place in which additions for the configuration functionality for a new DA method need to be done.
When adding a DA method, a line for the corresponding method-specific routine has to be inserted to each of the interface routines in PDAF_utils_filters.F90. Further, one has to add to PDAF_da.F90 a line for the DA method declaring its name in the form PDAF_DA_X and a correspondig index.
|
Internal structure of a DA method's code
Modules of a DA method
Each DA method in PDAF consist of 5 standard modules. These are
PDAF_X | This module declares variables specific to the DA method, e.g. the variable for the ensemble inflation. Further, it contains the different subroutines called by the routines in PDAF_utils_filters for the configuration of the DA method.
|
PDAFassimilate_X | This module contains the assimilation interface routines. Usually these are PDAF_assimilate_X and PDAF_assim_offline_X.
|
PDAFput_state_X | This module contains the routine PDAF_put_state_X. This routine controls the ensmeble integrations for the flexible parallelization variant. It exists as a spearate routine for backward-compatibility with PDAF2.
|
PDAF_X_update | This module contains the main routine PDAFX_update for the analysis update. This controls the actual update, e.g. by performing a local analysis loop for domain-localized filter methods.
|
PDAF_X_analysis | This module contains the routine that computes and applies the actual ensemble analysis increment. |
Note on naming modules and subroutines: Fortran does not allow that the name of a module is idential to the name of a subroutine contained in the module. We we handle this issue with using or omitting understores, e.g. the module PDAFassimilate_X contains the subroutine PDAF_assimilate_X.
Calls structure for analysis step
The routine PDAFomi_assimilate_local, for local ensemble filters and smoothers, or analogously the other available PDAF_omi_assimilate routines, like for global ensemble filters and smoothers (PDAFomi_assimilate_global) or the different variants of 3D-Var (PDAFomi_assimilate_*3dvar*) are called directly from the model code. These are used for the fully-parallel implementation variant. For the flexible-parallelization variant the analogous routines PDAFomi_put_state_* are provided. These are generic routines which internally call the method-specific routine (PDAF_assimilate_X or PDAF_put_state_X) according to the chosen filter which then calls the actual update routine (PDAF_X_update). This call structure is explained in Figure 2.
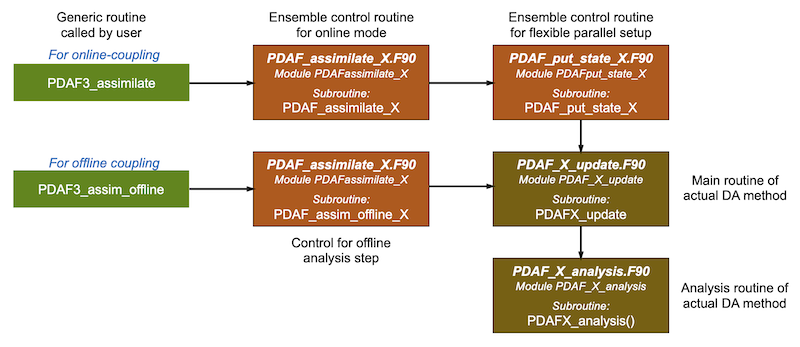
Figure 2: Internal call structure for the analysis step. The universal interface routines PDAF3_assimilate and PDAF3_Assim_offline call a corresponding specific routine of the DA method. These specific routines are PDAF_assimilate_X and PDAF_assim_offline_X which are members of the module PDAFassimilate_X. PDAF_assimilate_X calls PDAF_put_state_X. These two routines together control the online coupled mode, while PDAF_assim_offline_X controls the offline coupled mode. The actual analysis update is performed by the routines, PDAFX_update and PDAFX_analysis, each in their own module.
The universal routine PDAF3_assimilatel as example) calls the method-specific routine. Here PDAF_assimilate_X is the routine that controls the ensemble run in case of the fully-parallel implementation. The routine PDAF_put_state_X is used for both the fully-parallel and flexible parallelization variants. For the flexible parallelization one uses PDAFomi_put_state_local which directly calls PDAF_put_state_X, while for the fully parallel variant this routine is called by PDAF_assimilate_X. PDAF_put_state_X controls the ensemble for the case that multiple ensmeble states are propagated by a single model task and collects the ensemble from the ensemble tasks before the assimilate update and distributes the ensemble to the model tasks afterwards. The actual main routine for the DA method is PDAF_X_update which is called by PDAF_put_state_X. The routines on the right without 'omi' in their name are those with the full interface so that they are usable without OMI.
See the section on filter-specific routines for a detailed description for PDAF_assimilate_X and PDAF_put_state_X.
Internal dimensions
PDAF internally stores the dimensions of the assimilation system. The dimensions are declared in the Fortran module PDAF_mod_filter. Important are the following dimensions:
dim_p | The size of the state vector (with parallelization the size of the local state vector for the current process) |
dim_ens | The overall size of the ensemble |
dim_ens_l | If the ensemble integration is distributed over several ensemble tasks, this variable stores the size of the sub-ensemble handled by the current process. (dim_ens_l equals dim_ens if no parallelization or if only a single model task is used.) For the fully parallel implementation it is din_ens_l=1. Note that this variable is only used in the ensemble handling of PDAF, but not in the DA update.
|
rank | The maximum rank of the ensemble covariance matrix. In almost all cases, it is dim_ens-1. Used in error-subspace filters, (L)ESTKF and (L)SEIK
|
Internal arrays
Several internal arrays are allocated when PDAF is initialized. These arrays are declared in PDAF_mod_filter. They are allocated in PDAF_X_alloc (see below for details) and remain allocated throughout the assimilation process.
For the processes that computes the analysis (those with filterpe=.true.) the following arrays are defined:
| Array | Dimension | Comment |
state | dim_p | State vector. Used in all filters. Inside the filter code, it's usually called state_p
|
eofV | dim_p x dim_ens | Ensemble array. Used in all filters. Inside some filters the name is ens_p
|
eofU | dim_ens-1 x dim_ens-1 (SEEK, SEIK, ESTKF)dim_ens x dim_ens (ETKF) | Eigenvalue matrix U from P=VUVT (SEEK, SEIK) or transform matrix A (ETKF, ESTKF). Not used in EnKF. In some routines the matrix is called Uinv to indicate the inverse
|
state_inc | dim_p | state increment vector. Only allocated if incremental analysis updates are used |
For the processes that only compute model forecasts but are not involved in the analysis step (i.e. filterpe=.false.), only one array is defined:
| Array | Dimension | Comment |
eofV | dim_p x dim_ens_l | Ensemble array on non-filter processes. Used in all filters. |
Filter-specific routines
When a filter algorithm is added, the following filter routines have to be implemented and inserted to each interface routines described above.
PDAF_X_initPDAF_X_allocPDAF_X_options(optional)PDAF_X_memtime(optional)
The routines are very similar for all DA methods.
In addition, one has to implement the routines
PDAF_put_state_XandPDAF_assimilate_X
As an example we recommend to see e.g. the routines for the ETKF or LETKF.
We recommend to base on the routines of an existing filter, as most of the routines can be easily adapted to a new filter method.
PDAF_X_init
The routine PDAF_X_init performs the initialization of filter-specific parameters. In addition, it prints information about the configuration.
The interface is as follows:
SUBROUTINE PDAF_X_init(subtype, param_int, dim_pint, param_real, dim_preal, &
ensemblefilter, fixedbasis, verbose, outflag)
INTEGER, INTENT(inout) :: subtype ! Sub-type of filter
INTEGER, INTENT(in) :: dim_pint ! Number of parameters in param_int
INTEGER, INTENT(inout) :: param_int(dim_pint) ! The array of integer parameters
INTEGER, INTENT(in) :: dim_preal ! Number of parameters in param_real
REAL, INTENT(inout) :: param_real(dim_preal) ! The array of real parameters
LOGICAL, INTENT(out) :: ensemblefilter ! Flag, whether the filter is an ensemble filter or a mode-based filter
LOGICAL, INTENT(out) :: fixedbasis ! Flag, whether the chosen `subtype` uses a fixed ensemble (only the ensemble mean is integrated by the model)
INTEGER, INTENT(in) :: verbose ! Control screen output
INTEGER, INTENT(inout) :: outflag ! Status flag
The routine has to perform the following operations:
- initialize the PDAF-internal parameter variables specific for the DA method from the provided values of
subtype,param_int, andparam_real. - set the logical flags
ensemblefilterandfixedbasis.
The existing implementations also include some screen output about the configuration.
Please note:
- The routine should check, whether the provided value of
subtypeis a valid choice. If this is not the case, the error flag should be set to 2. - Only parameters from
param_intandparam_realup to the valuedim_pintanddim_preal, respectively, should be considered in the initialization. The arrays may be bigger, but the user defines which parameters are to be used be setting the values ofdim_pintanddim_preal. - The error flag
outflagis initially set to 0. - The internal parameters are declared in the Fortran module
PDAF_mod_filter. If a new filter algorithm requires additional parameters, their declaration should be added to the module. Alternatively, one can introduce a new module the holds the parameters specific to the new DA method.
PDAF_X_alloc
The routine PDAF_X_alloc allocates arrays for the data assimilation. These are the arrays that are used in the forecasting and hence need to be persistently allocated, like the ensemble array and a state vector. The success of the allocation is checked.
The interface is as follows:
SUBROUTINE PDAF_X_alloc(subtype, outflag) INTEGER, INTENT(in) :: subtype ! Sub-type of filter INTEGER, INTENT(out):: outflag ! Status flag
All arrays that need to be allocated are declared in the Fortran module PDAF_mod_filter. Here, also the shapes of the arrays are declared. For the allocation of arrays, one has to distinguish between processes that compute the analysis step (filterpe=.true.) and those that only participate in the ensemble forecast (filterpe=.false.).
For the processes that compute the analysis (those with filterpe=.true.) it is mandatory to allocate the following two arrays:
state: The state vector of sizedim_p.eofV: This is the ensemble matrix in all ensemble-based filters. For SEEK it is the matrix holding eigenvectors.eofVhas size (dim_p,dim_ens).
Depending on the filter algorithm some of the following arrays also need to be allocated:
eofU: This is the eigenvalue matrix U used in the SEIK and SEEK filters and the transform matrix of ESTKF (here, its size is (rank,rank)). For ETKF, it is the transform matrix A of size (dim_ens,dim_ens). The array should always be allocated. For method that don't use such transform matrix, for example the EnKF, it can be allocated with size (1,1).state_inc: The increment to the state vector computed in the analysis step. It only needs to be allocated in this routine, if incremental analysis updating is implemented. Otherwise, it is sufficient to allocate and deallocatestate_incin the routine performing the analysis step. The size ofstate_incisdim_p.bias: If the filter algorithm is implemented with bias correction, the vectorbiaswith sizedim_bias_pis allocated.
Processes that only participate in the computation of the ensemble forecast, but are not involved in computing the analysis step (those with filterpe=.false.), operate only on a sub-ensemble. Accordingly, an ensemble array for this sub-ensemble has to be allocated. This is:
eofV: This is the ensemble matrix in all ensemble-based filters. For SEEK it is the matrix holding eigenvectors. For the processes withfilterpe=.false.,eofVhas size (dim_p,dim_ens_l).
PDAF_X_options
The optional routine PDAF_X_options displays information on the available options for the filter algorithm.
The routine has no arguments. Thus the interface is as follows:
SUBROUTINE PDAF_X_options()
The following display is recommended:
- Available subtypes (At least '0' for standard implementation)
- Parameters used from the parameter arrays
param_intandparam_real.
PDAF_X_memtime
The optional routine PDAF_X_memtime displays information about allocated memory and the execution time of different parts of the filter algorithm.
The interface is as follows:
SUBROUTINE PDAF_X_memtime(printtype)
INTEGER, INTENT(in) :: printtype ! Type of screen output:
! (1) general timings
! (3) timing information for call-back routines and PDAF-internal operations
! (4) second-level timing information
! (5) very detailed third-level timing information
! (10) process-local allocated memory
! (11) globally allocated memory
The timing operations are implemented using the module PDAF_timer, which provides the function PDAF_timeit. Memory allocation is computed using PDAF_memcount, which is provided by the module PDAF_memcounting.
PDAF_assimilate_X / PDAF_put_state_X
These routines are called by PDAFomi_assimilate_* or PDAFomi_put_state_*, which are directly inserted into the model code, if the online mode of PDAF is used together with PDAF-OMI. The description of the implementation of the analysis step in the Implementation Guide explains the interface for the algorithms that are included in the PDAF package. PDAFomi_assimilate_* are interface routines for PDAF-OMI and call the PDAF_assimilate_X routines as described above. (Likewise the routines PDAFomi_put_state_* call PDAF_put_state_X.)
As described before, PDAF_assimilate_X calls PDAF_put_state_X which then calls the actual update routine of the DA method. Here. PDAF_assimilate_X mainly passes its arguments over to PDAF_put_state_X, but it also controls the ensemble run by counting the number of time steps.
Apart from the usual integer status flag, the interface of the routines contains the names of the user-supplied call-back 2routines that are required for the analysis step. Usually, the minimum set of routines are:
U_collect_state: The routine that writes model fields into the state vector, i.e. a single column of the ensemble state arrayU_init_dim_obs: The routine that determines the size of the observation vectorU_obs_op: The routine that contains the implementation of the observation operatorU_init_obs: The routine that provdes the vector of observationsU_prepoststep: The pre- and post-step routine in which the forecast and analysis ensembles can be analyzed or modified.
PDAF_assimilate_X uses in addition
U_distribute_state: The routine that fills the field arrays of a model from the state vector, i.e. a single column of the ensemble state arrayU_next_observation: The routine that specified the number of time steps until the next DA analysis update.
Further routines can be added and depend on the requirements of the filter algorithm. Common is a routine that involves the observation error covariance matrix. For example, the (L)ETKF and (L)ESTKF methods use the routine
U_prodRinvAwhich has to multiply some temporary matrix of the filter method with the inverse observation error covariance matrix.
When one plans to implement a new filter, we recommend to check whether the filter is compatible with the existing local or global filters, in particular whether it uses the same call-back routines. In this case, the new filter can be added into the existing routines (either PDAFomi_assimilate_global/PDAFomi_put_state_global for a global DA method or likewise PDAFomi_assimilate_local/PDAFomi_put_state_local for a DA method using domain localization).
The routine PDAF_assimilate_X and PDAF_put_state_X provide the actual infrastructure to manage the ensemble forecasting. Here PDAF_assimilate_X is used for the fully parallel setup and counts the time steps and finally calls PDAF_put_state_X.
The routines PDAF_put_state_X prepare for the actual analysis step, that is called inside these routines as a separate routine PDAF_X_update. PDAF_put_state_X also performs the ensemble management for the flexible parallelization variant in which more than one ensemble state can be integrated by a model task.
The operations implemented in PDAF_put_state_X are:
- Write model fields back into the ensemble array (by calling
U_collect_state) - Increment the counter for the integrated ensemble members (named
counterand provided by the modulePDAF_mod_filter. - Check, if the ensemble integration is completed (in that case, it is
member = local_dim_ens + 1). If not, exitPDAF_put_state_X. - When the ensemble integration is completed, the following operations are required:
- If more than one model task is used: Collect the sub-ensembles from all model tasks onto the processes that perform the analysis step. This operation is done by the subroutine
PDAF_gather_ens. - Call the routine that computes the analysis step for the chosen filter algorithm (typically named
PDAF_X_update). - Reset the control variables for the ensemble forecast (
initevol=1,member=1,step=step_obs+1).
- If more than one model task is used: Collect the sub-ensembles from all model tasks onto the processes that perform the analysis step. This operation is done by the subroutine
In general, the PDAF_put_state routines of all ensemble-based filters have the same structure. For the implementation of a new filter we recommend to base on an existing routine, e.g. that of for the ETKF. Thus, one copies the routine to a new name. Then, one adapts the interface for the required user-supplied routines of the new DA method. In addition, the call of the routine PDAF_X_update holding the DA analysis update step has to be revised (name of the routine, required user-supplied routines).
The routine PDAF_assimilate_X is mainy an interface routine to PDAF_put_state_X. It counts the time steps and calls PDAF_put_state_X when the forecast phase is complete. Thus, to implement a new filter, one can copy a routine and apart form adapting the name (X) and the interface specifying the call-back routines, there should be no need for changes.
Analysis update step PDAF_X_update
The actual DA analysis update is computed in the routine PDAF_X_update. The routine is provided with the names of the call-back routines, the arrays as well of the relevant dimensions as describe above.
The structure of the operations in PDAF_X_update can be designed freely when implementing a new DA method. The existing methods follow the recommended structure in which PDAF_X_update only performes preparations for the actual analysis update and then calls PDAF_X_analysis for the computation of the actual update. Here, the operations are different for global and local methods.
Global Methods
For global methods, e.g. ESTKF, ETKF, EnKF, NETF, the general structure of PDAF_X_update is:
IF (fixed ensemble, i.e. subtype==2 or subtype==3) THEN
Add central state and ensemble perturbations
ENDIF
CALL U_prepoststep # Perform pre/poststep before analysis update
CALL PDAF_X_analysis # Compute global analysis update of ensemble
CALL PDAF_smoother # Compute smoother update (Only for filters for which a smoother is implemented)
CALL U_prepoststep # Perform pre/poststep after analysis update
You will find this structure e.g. in PDAF_ESTKF_update.F90. This structure is also valid for the LEnKF. The actual implementation, e.g. in PDAF_ESTKF_update.F90 contains additional functionality e.g. calls for timers and normal screen output or debug outputs. Also there can be several different calls PDAF_X_analysis depending on the subtype of a filter.
This general structure should be widely usable. Thus, when implementing a new global ensemble method one could copy an existing file and adapt it for the new method. The main functionality of the new DA method would then be implemented in PDAF_X_analysis.
Local Methods
For domain-localized Methods, e.g. LESTKF, LETKF or LNETF the general structure of PDAF_X_update is different from the global methods because the local analysis loop is contained in PDAF_X_update and only a local analysis routine is called. The general structure is:
IF (fixed ensemble, i.e. subtype==2 or subtype==3) THEN
Add central state and ensemble perturbations
ENDIF
CALL U_prepoststep # Perform pre/poststep before analysis update
CALL U_init_n_domains # Determine number of local analysis domains (n_domains_p)
# --- Perform global operations on observations and ensemble
CALL U_init_dim_obs # Determine number of observations (PDAF-OMI routine init_dim_obs_pdafomi)
DO i = 1, dim_ens
CALL U_obs_op # Apply observation operator to all ensemble state (PDAF-OMI routine obs_op_pdafomi)
ENDDO
Compute mean forecast state (state_p)
Compute mean observed forecast state (HXbar_f)
# --- End of global operations
# --- Perform local analysis loop
DO i = 1, n_domains_p
CALL U_init_dim_l # Set size of local state vector
CALL U_init_dim_obs_l # Set number of observations within radius (PDAF-OMI routine init_dim_obs_l_pdafomi)
DO i = 1, dim_ens
CALL U_g2l_state # Get local state vector for all ensemble states
ENDDO
CALL PDAF_X_analysis # Compute local analysis update of ensemble
DO i = 1, dim_ens
CALL U_l2g_state # Update global state vector for all ensemble states
ENDDO
CALL U_l2g_state # Update global state vector for ensemble mean state
CALL PDAF_smoother_local # Only for filters for which a smoother is implemented
ENDDO
# --- End of local analysis loop
CALL U_prepoststep # Perform pre/poststep after analysis update
You will find this structure e.g. in PDAF_LESTKF_update.F90. This structure is not used for the LEnKF because this filter doe snot use a local analysis loop. The actual implementation, e.g. in PDAF_LESTKF_update.F90 contains additional functionality e.g. calls for timers and normal screen output or debug outputs. Also there can be different calls PDAF_X_analysis depending on the subtype of a filter.
This general structure should be widely usable for DA methods that use domain localization. Accordingly, when implementing a new local ensemble method one could copy an existing file and adapt it for the new method. The main functionality of the new DA method would then be implemented in PDAF_X_analysis.
